
An important paper on the genetics of keratoconus offered a detailed look at the various genetic analysis techniques and whether they could be successfully applied to finding a genetic basis for KC.
An important paper on the genetics of keratoconus was recently published in the journal Ophthalmic Genetics. Scientists based at the University of Antwerp (Belgium) offered a detailed look at the various genetic analysis techniques in use, and whether they could be successfully applied to finding a genetic basis for KC.
The authors wrote that KC appears to be a disease continuum. It appears to have a genetic contribution, combined with environmental factors. Sometimes KC shows the characteristics of monogenic disease; in most cases it appears to be a complex disease.
What does that mean?
In a monogenic disease, a single error in a single gene causes the disease. There are reports of large families with an extraordinary high rate of KC. In these cases, researchers believe there may be a single gene abnormality responsible, and the mode of inheritance is described as autosomal dominant inheritance with reduced penetrance. This means that in these affected families, some individuals who carry the gene mutation may show clinical signs of KC, while others may not exhibit any symptoms but would still carry the “KC gene”. However, a single gene mutation only explains a small percentage of keratoconus cases.
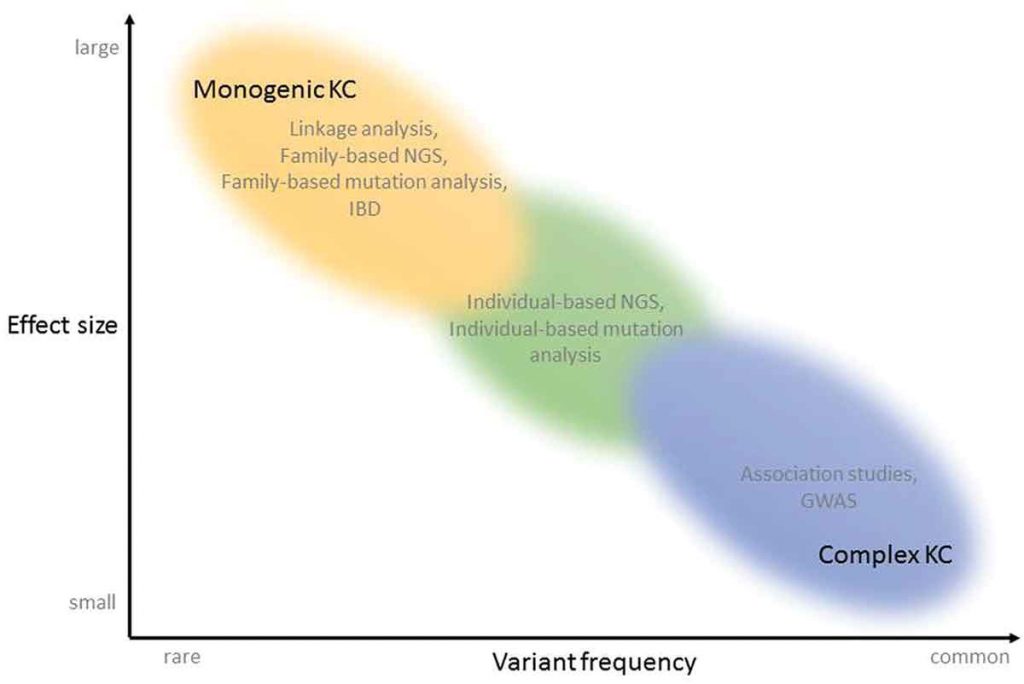
Figure 1. Techniques that have been used for the identification of genetic variants responsible for KC, in function of variant frequency and effect size that can be detected with the techniques. KC can present as a monogenic disease (top left of the graph), with one large effect, rare, mutation per patient. Linkage analysis, Identity-by-descent, Family based Next-Generation Sequencing (NGS) and Family based mutation analysis have been applied to identify the disease-causing mutations in patients with the monogenic form of KC. The complex forms of KC are located lower and more to the right of the graph, in the area of more common variants with smaller effect size. Genome-wide association studies and association studies investigate common variants. The variants located in the middle of the continuum, between rare and common and between small and large effect sizes, can be identified using individual-based NGS and mutation analysis. KC: keratoconus, NGS: Next-Generation Sequencing, IBD: Identity-by-descent, GWAS: Genome Wide Association Study.
In a complex disease, each affected individual has a number of small genetic variations in their DNA (for example, genes that impact corneal thinning, or collagen stiffness, or even corneal curvature).
Alone the variations may not be enough to cause KC, but the cumulative effect of the variations, along with environmental factors (such as eye rubbing or allergies), result in KC.
If KC is a continuum, and there are individuals whose disease is the result of a single gene mutation, there are many more individuals at the other end of the spectrum whose disease is caused by environmental factors in combination with a large number of small effect genetic risk factors.
The authors suggest that future research explore the interplay between smaller mutations, as well as the interplay between these small genetic variations and environmental factors to clarify their link to KC. They also suggested that areas of the human genome that are not well understood may provide answers about true disease-causing KC genes.
Resource:
1. Valgaeren H, Koppen C, Van Camp G, “A new perspective on the genetics of keratoconus: why have we not been more successful? Ophthalmic Genet 2017 Nov 7:1-17.

